PyDI: Python Data Integration package |
||
SourceForge project
Download PyDI
API Documentation
Browse source

|
PyDI is a lightweight, 100% Python, federated data integration system. Users may create custom schemas for disparate sources, query and expand results across sources to find related data. The primary usage for PyDI is to facilitate graph-based data mining of biomedical repositories (or any set of repositories that can be interlinked but are inherently fractured and disparate).
Supported features: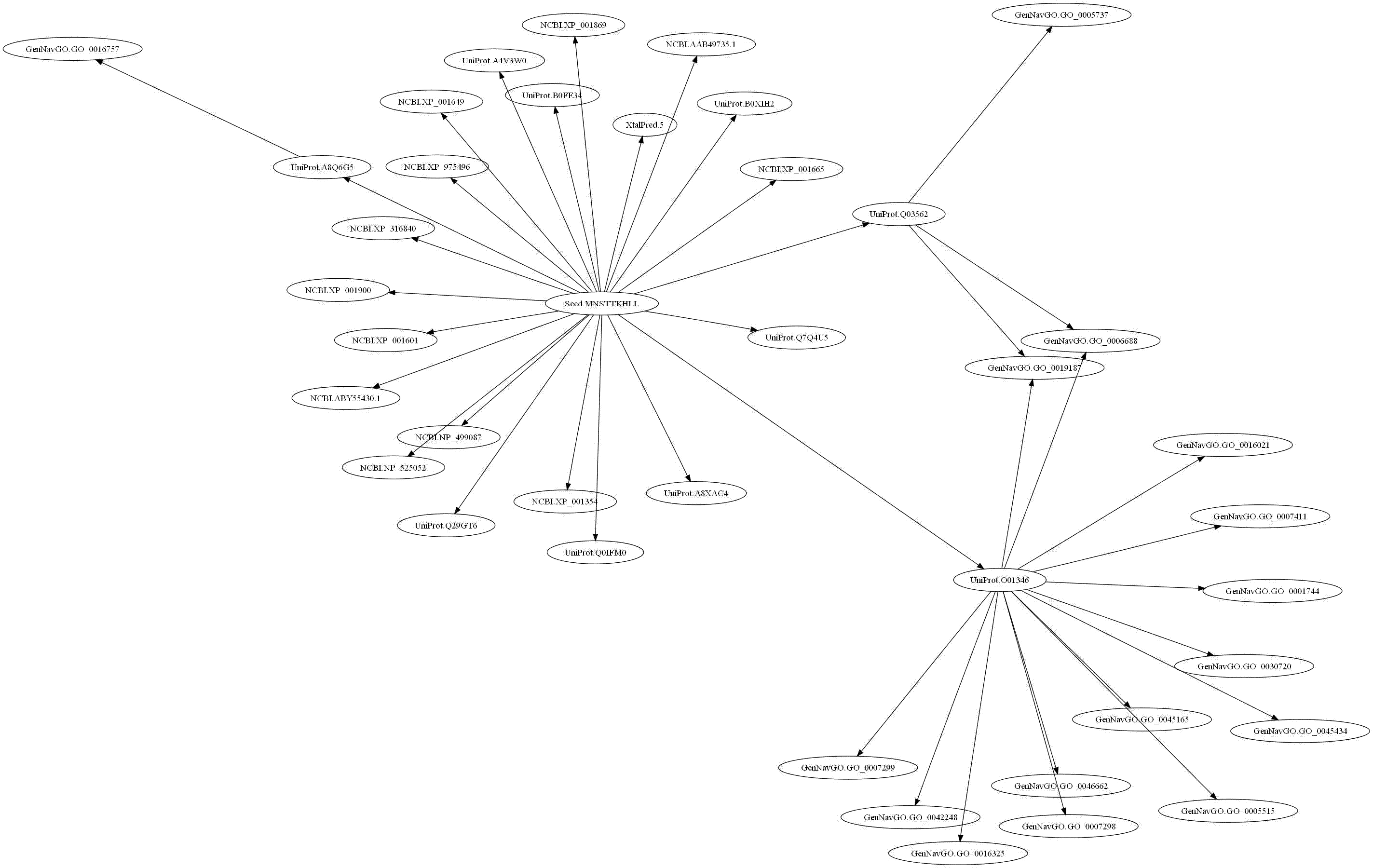
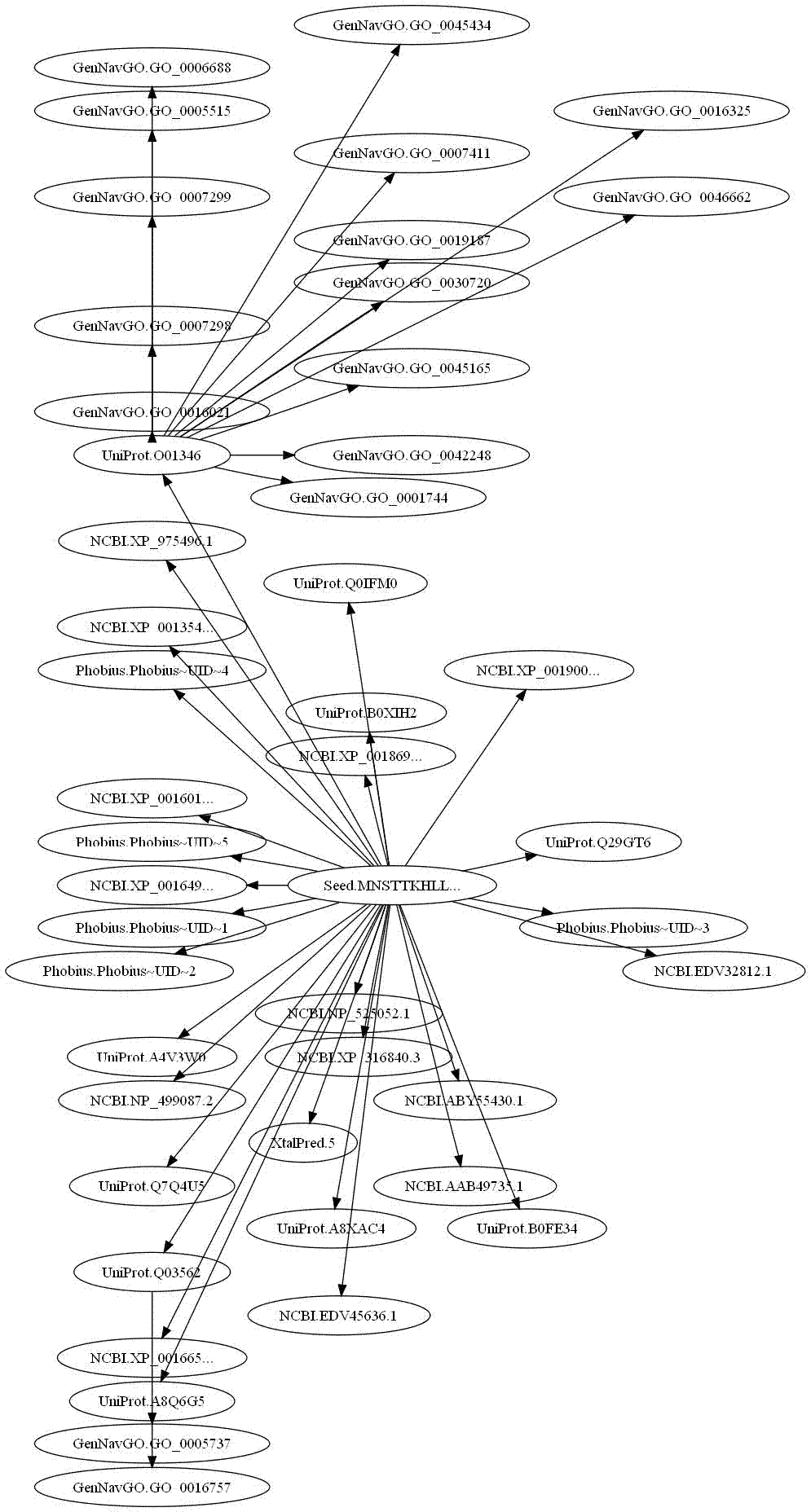
Example usage:Querying a sequence of unknown function across multiple biologic data sources:
# Instantiate a browser with the preferred schema, and with
# source interfaces in a module called 'generators'.
>>> browser = BrowserEngine("biological_schema.txt","generators")
# Wrap a graph around the engine, to explore the data later
# using graph-theoretic methods.
>>> G = NXBrowserGraph(browser)
# Seed a protein query (the source 'UserQuery' and entity 'Protein'
# are definable in the schema.
>>> browser.seed("UserQuery","Protein","Sequence","MNSTTKHLLH....")
# Now enlarge the seeded result up to two expansions
>>> auto_expand_browser(browser,2)
->DB:MNSTTKHLLH... is loaded into database UserQuery.
->QUERY: NCBI.MNSTTKHLLH...
->QUERY: UniProt.MNSTTKHLLH...
...
...
...
# Generate an image of the results graph.
>>> generate_graph_jpg(browser,"unknown.jpg")
>>>
|
|
|
Copyright © 2008 Eithon Cadag |
||